Maybe it is just confirmation bias, but there seems to have been an upswing in the number of cool papers on the role of intraspecific variation in ecology. For example, three new papers highlight the importance of variation among individuals for topics ranging from conservation, coexistence, and community responses to changing environments. All are worth a deeper read.
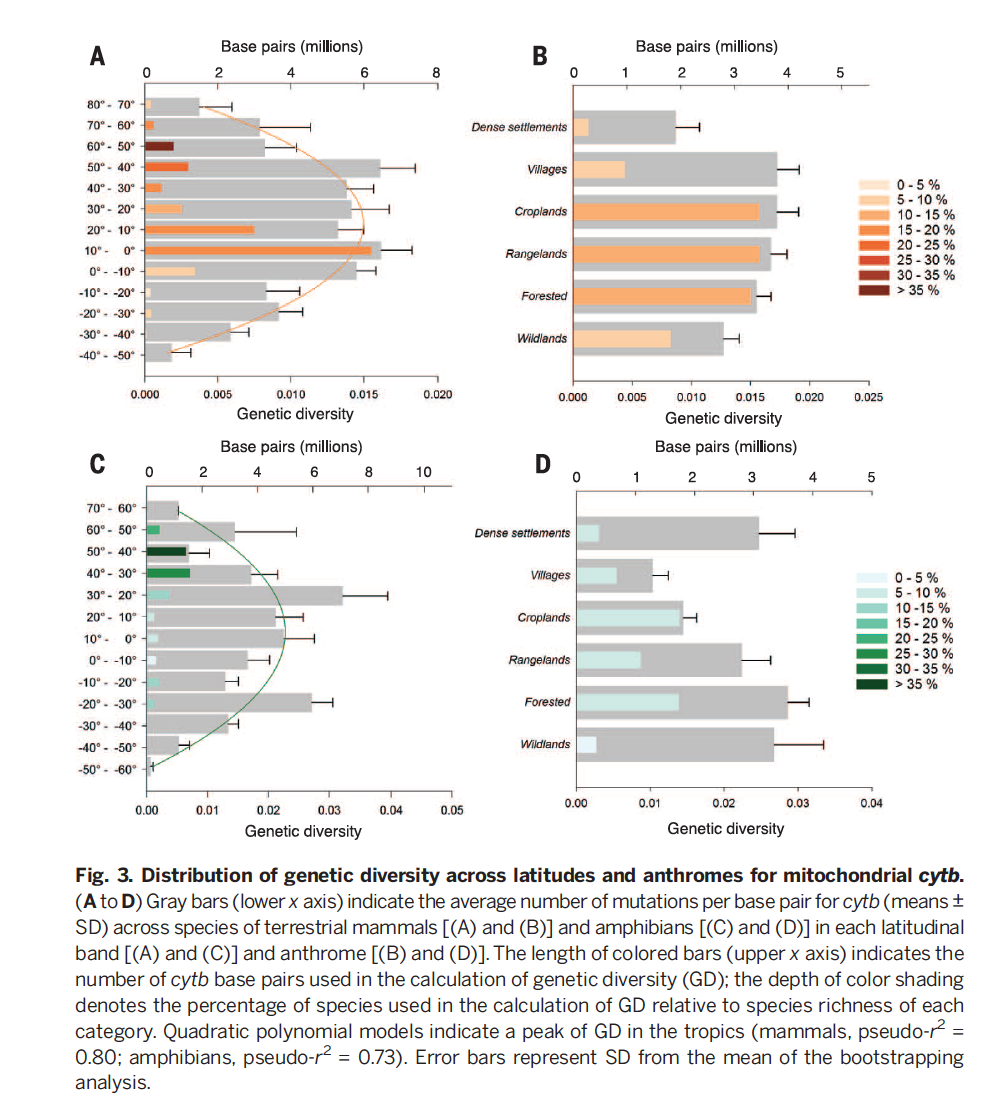
‘An Anthropocene map of genetic diversity’ asks how intraspecific variation is distributed globally, a simple but important question. Genetic diversity in a species is an important predictor of their ability to adapt to changing environments. For many species, however, as their populations decline in size, become fragmented, or experience strong selection related to human activities, genetic diversity may be in decline. Quantifying a baseline for global genetic diversity is an important goal. Further, with the rise of ‘big data’ (as people love to brand it) it is now an accessible one: there are now millions of genetic sequences in GenBank and associated GPS coordinates.
‘An Anthropocene map of genetic diversity’ asks how intraspecific variation is distributed globally, a simple but important question. Genetic diversity in a species is an important predictor of their ability to adapt to changing environments. For many species, however, as their populations decline in size, become fragmented, or experience strong selection related to human activities, genetic diversity may be in decline. Quantifying a baseline for global genetic diversity is an important goal. Further, with the rise of ‘big data’ (as people love to brand it) it is now an accessible one: there are now millions of genetic sequences in GenBank and associated GPS coordinates.
Many of the global patterns in genetic diversity agree with those seen for other forms of diversity: for example, some of the highest levels are observed in the tropical Andes and Amazonia, and there is a peak in the mid-latitudes and human presence seems to decrease genetic diversity.
 |
| From Miraldo et al. (2016): Map of uncertainty. Areas in green represent high sequence availability and taxonomic coverage (of all species known to be present in a cell). All other colors represent areas lacking important data. |
The resulting data set represents ~ 5000 species, so naturally the rarest species and the least charismatic are underrepresented. The authors identify this global distribution of ignorance, highlighting just how small our big data still is.
Miraldo, Andreia, et al. "An Anthropocene map of genetic diversity." Science353.6307 (2016): 1532-1535.
In ‘How variation between individuals affects species coexistence’, Simon Hart et al. do the much needed work to answer the question of how intraspecific variation fits into coexistence theory. Their results reinforce the suggestion that in general, intraspecific variation should making coexistence more difficult, since it increases the dominance of superior competitors, and reduces species' niche differentiation. (Note this is a contrast to the argument Jim Clark has made with individual trees, eg. Clark 2010)
Hart, Simon P., Sebastian J. Schreiber, and Jonathan M. Levine. "How variation between individuals affects species coexistence." Ecology letters (2016).
The topic of evolutionary rescue is an interesting, highlighting (see work from Andy Gonzalez and Graham Bell for more details) the ability of populations to adapt to stressors and changing environments, provided enough underlying additive genetic variation and time is available. It has been suggested that phenotypic plasticity can reduce the chance of evolutionary rescue, since it reduces selection on genetic traits. Alternatively, by increasing survival time following environmental change, it may aid evolutionary rescue. Ashander et al. use a theoretical approach to explore how plasticity interacts with a change in environmental conditions (mean and predictability/autocorrelation) to affect extinction risk (and so the chance of evolutionary rescue). Their results provide insight into how the predictability of new environments, through an affect on stochasticity, in turn changes extinction risk and rescue.