Community ecology is difficult in part because it is so multi-dimensional: communities include possibly hundreds of species present, and in addition the niches of each of those species are multi-dimensional. Functional or trait-based approaches to ecology in particular have been presented as a solution to this problem, since fewer traits (compared to the number of species) may be needed to capture or predict a community’s dynamics. But even functional ecology is multi-dimensional, and many traits are necessary to truly understand a given species or community. The question, when measuring traits to delineate a community is: how many traits are necessary to capture species’ responses to their biotic and/or abiotic environment? Too few and you limit your understanding, too many and your workload becomes unfeasible.
Plant communities in particular have been approached using a functional framework (they don't move, so trait measurements aren't so difficult), but the number and types of traits that are usually measured vary from study to study. Plant ecologists have defined functional groups for plants which are ecologically similar, identified particular (“functional”) traits as being important, including SLA, seed mass, or height, or taken a "more is more" approach to measurements. There are even approaches that capture several dimensions by identifying important axes (leaf-height-seed strategy, etc.). Which of these approaches is best is not clear. In a new review, Daniel Laughlin rather ambitiously attempts to answer how many (and which) traits plant ecologists should consider. He asks whether the multi-dimensional nature of ecological systems is a curse (there is too much complexity for us to ever capture), or a blessing (is there a limit on how much complexity actually matters for understanding these systems)? Can dimensionality help plant ecologists determine the number of traits they need to measure?
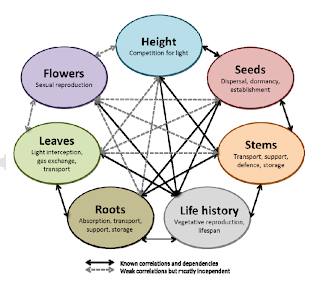 |
| From Laughlin 2013. The various trait axes (related to plant organs) important for plant function. |
Using 3 very large plant species-trait datasets (with 16-67(!) trait measures), Laughlin applies a variety of statistical methods to explore effective dimensionality reduction. He then estimates the intrinsic dimensionality (i.e. the number of dimensions necessary to capture the majority of the information in community structure) for the three datasets (figure below). The results were surprisingly consistent for each data set – even when 67 possible plant traits were available, the median intrinsic number of dimensions was only 4-6. While this is a reasonably low number, it's worth noting that the number of dimensions analyzed in the original papers using those datasets were too low (2-3 only).
 |
| From Laughlin 2013. The intrinsic number of traits/dimensions necessary to capture variation in community structure. |